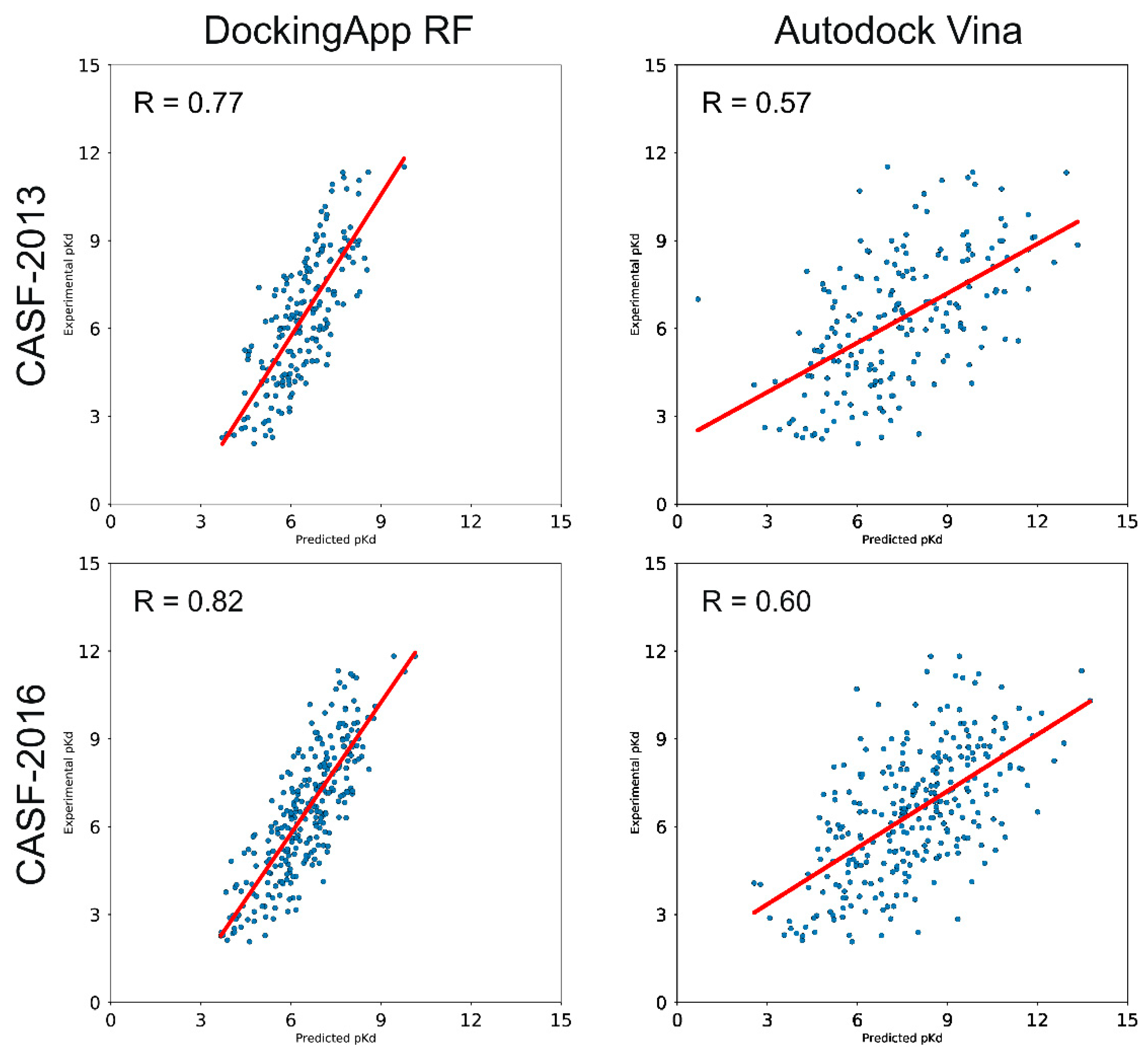
IJMS | Free Full-Text | DockingApp RF: A State-of-the-Art Novel Scoring Function for Molecular Docking in a User-Friendly Interface to AutoDock Vina

AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading - Trott - 2010 - Journal of Computational Chemistry - Wiley Online Library

Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock - ScienceDirect
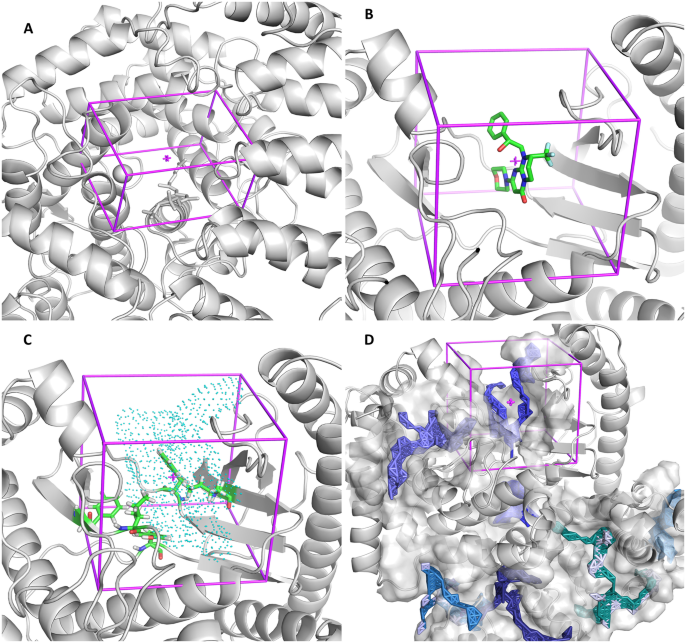
AMDock: a versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4 | Biology Direct | Full Text

Webina: An Open-Source Library and Web App that Runs AutoDock Vina Entirely in the Web Browser | bioRxiv

Molecular docking results employing AutoDock 4.2. Three dimensional... | Download Scientific Diagram
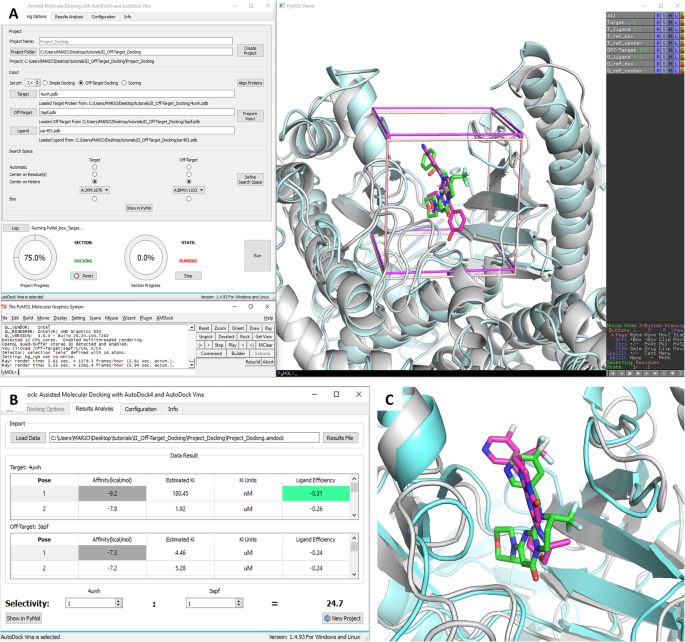
AMDock: a versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4 | Biology Direct | Full Text
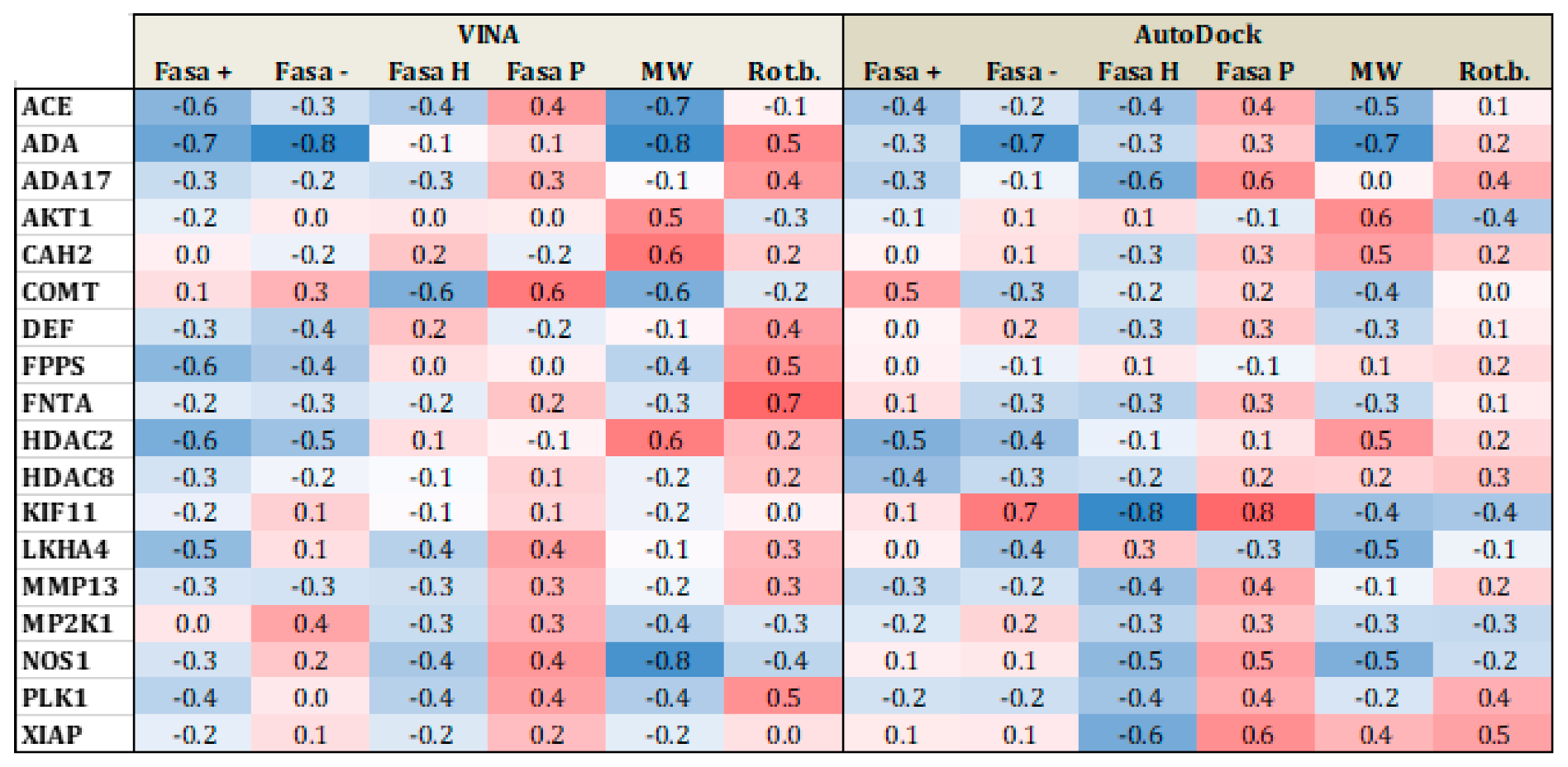
Applied Sciences | Free Full-Text | Comparing AutoDock and Vina in Ligand/Decoy Discrimination for Virtual Screening

IJMS | Free Full-Text | DockingApp RF: A State-of-the-Art Novel Scoring Function for Molecular Docking in a User-Friendly Interface to AutoDock Vina

Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark | Journal of Chemical Information and Modeling